1. Database Description
Rapid population growth poses a huge challenge to global agricultural production. Plant diseases caused by bacteria, fungi, oomycetes, and nematodes are one of the major factors limiting agricultural production and can cause yield losses of up to 100%. Understanding the plant-pathogen communication processes is an entry point for deciphering plant diseases. More and more pathogen effectors, as key roles in the plant-pathogen communication, were identified in pathogenic colonization, in which bioinformatics contributed in around 60% works. Therefore, bioinformatics is essential to accelerate the exploration of pathogen effectors.
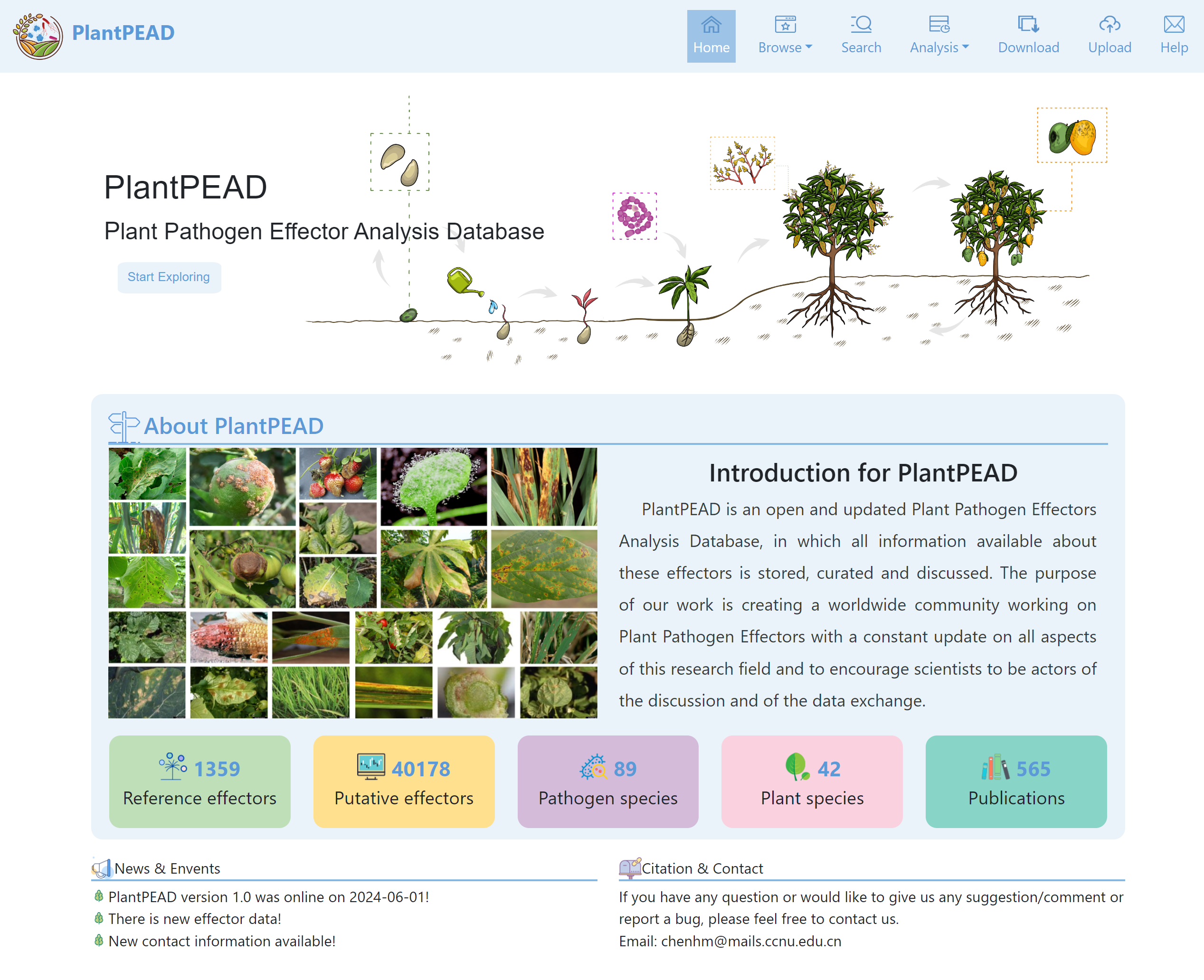
To this end, we established the Plant Pathogen Effector Analysis Database (PlantPEAD), which provides a comprehensive overview of effectors to facilitate multidisciplinary research. Our platform contains 1,359 experimentally identified and 40,178 orthologous identified pathogen effectors, of which more than 86% of experimentally identified effectors are derived from the literature. These effectors belong to 89 different pathogens infesting more than 40 plant species, including 426 plant-pathogen interactions, and are associated with useful biological information. In addition, we provide apoplastic and cytoplasmic localization of effectors. PlantPEAD is aimed at botanists, microbiologists, and bioinformaticians who may easily explore multidisciplinary research on combating plant diseases.
2. Database Access Instructions
Home Page
The homepage offers a comprehensive overview of our website. lt includes an introduction to the website, details about our services, information about us, and a contact section. We aim to give users a broad understanding of the webpage.
Browse Page
The browsing page offers several sections for easy navigation. This interface allows users to access pathogen effectors content from various perspectives, providing valuable information.
Disease page
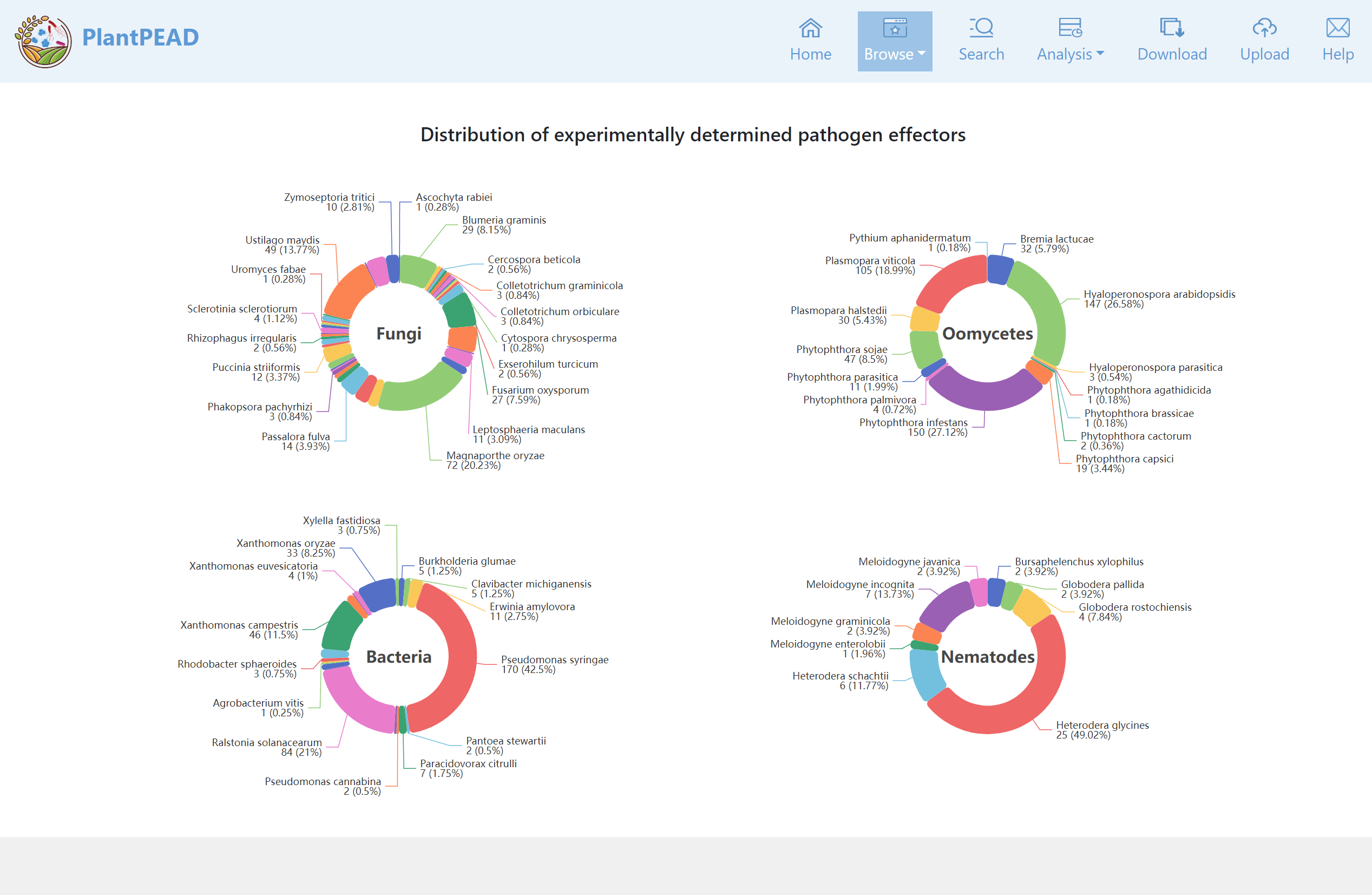
The disease page provides a categorization of experimentally determined pathogen effectors according to different disease categories.

Species page
On the species page, you can explore experimentally identified and orthologous identified pathogen effectors through different species.
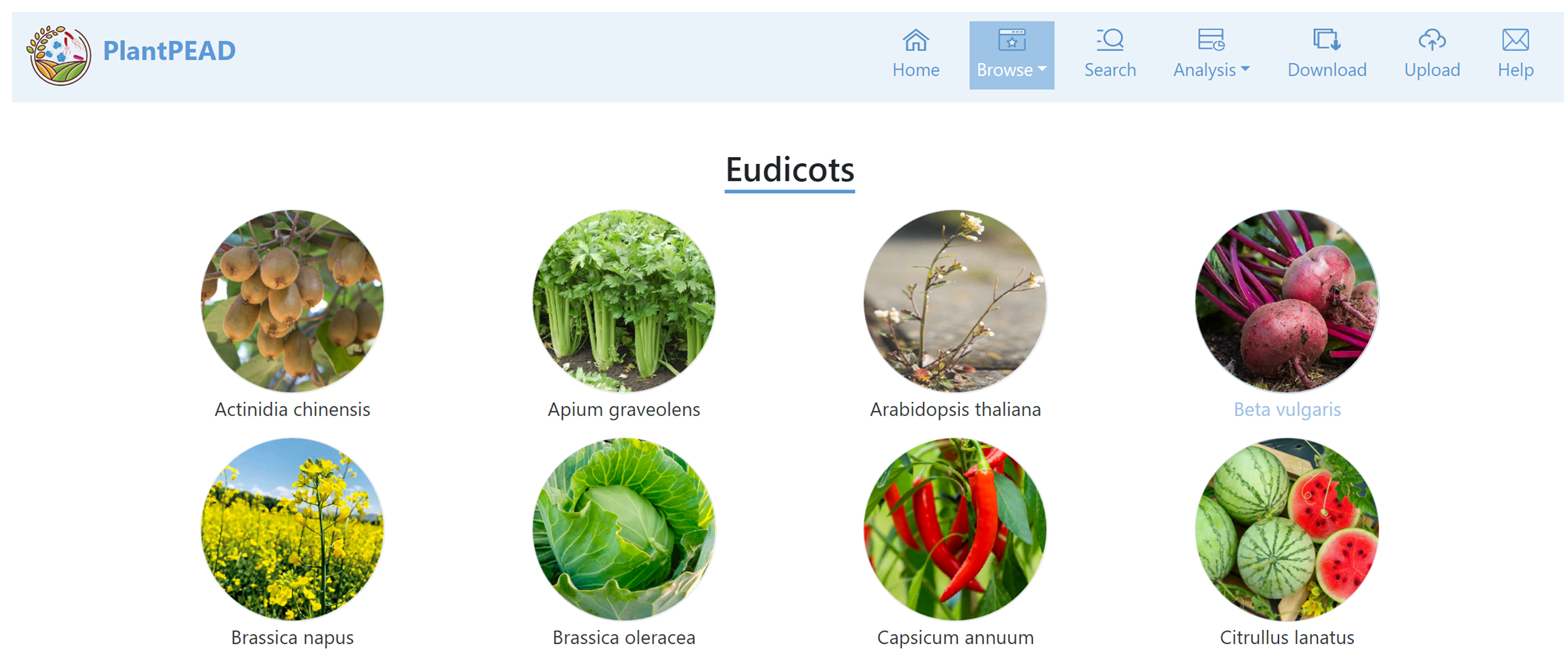
Plant page
In the plant page, you can explore 42 plant species categorized into eudicots, monocots, and others.
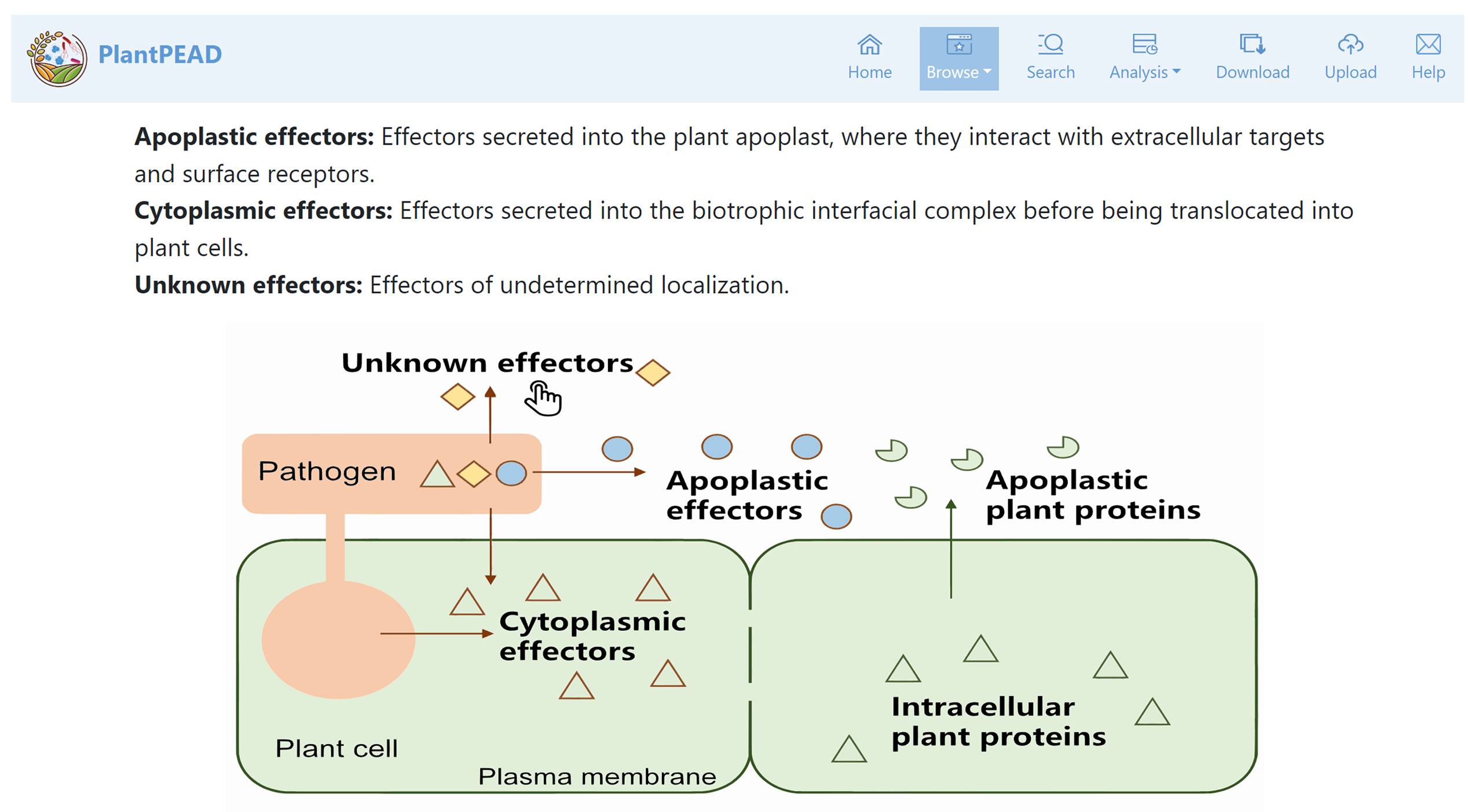
Localization page
In the localization page categorizes effectors based on apoplastic effectors, cytoplasmic effectors, and unknown effectors.
Pathogen Effector Information Page
After clicking through to this page from Browse, you will find pathogen effectors categorized by experimentally identified and orthologous identified. By clicking on a specific pathogen effector, you can access detailed descriptions related to that pathogen effector.
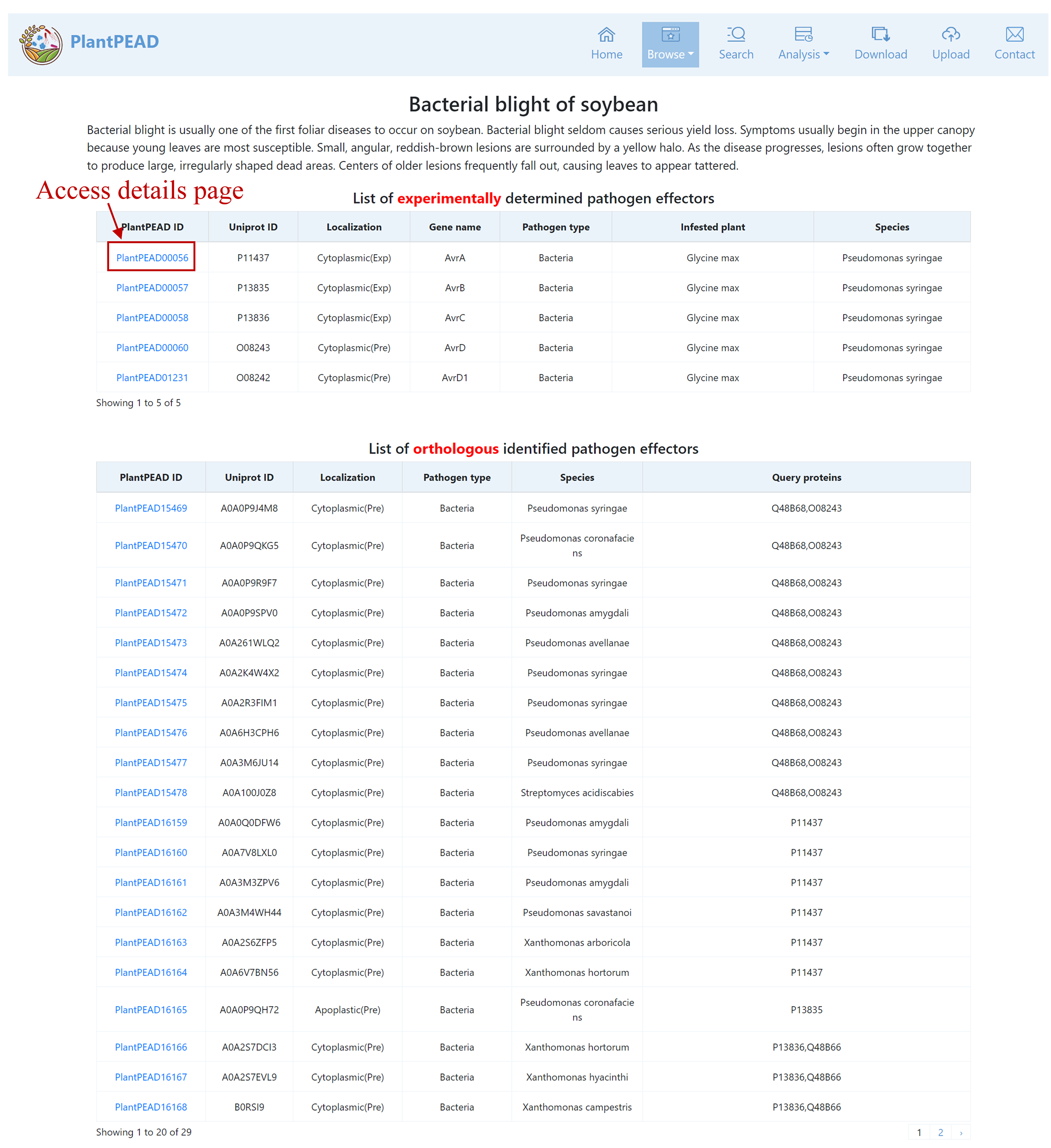
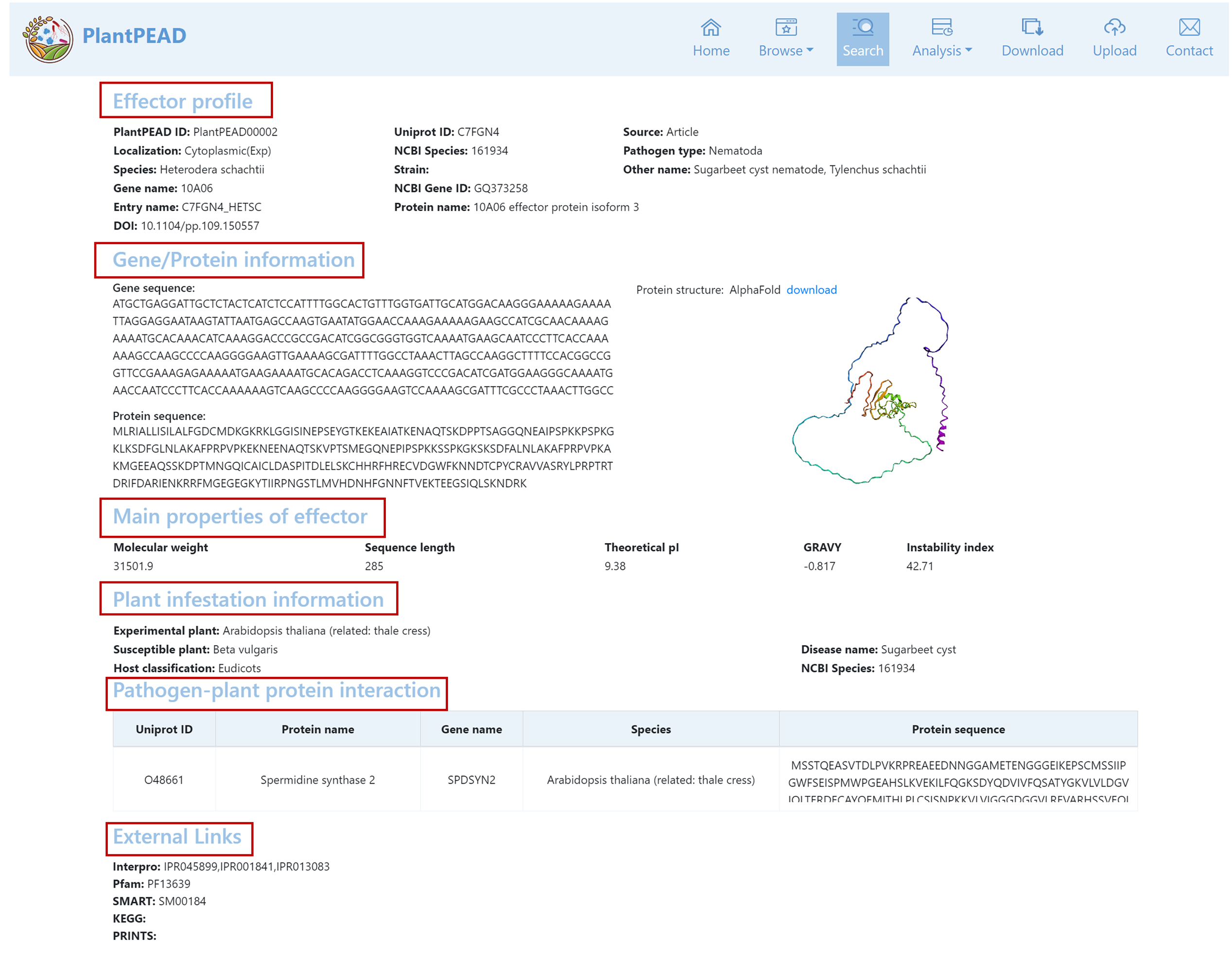
Pathogen Effector Details Page
The pathogen effector details page contains the effector profile, gene/protein information, physicochemical properties, plant infestation information, pathogen-plant protein interaction information, and functional annotation information.
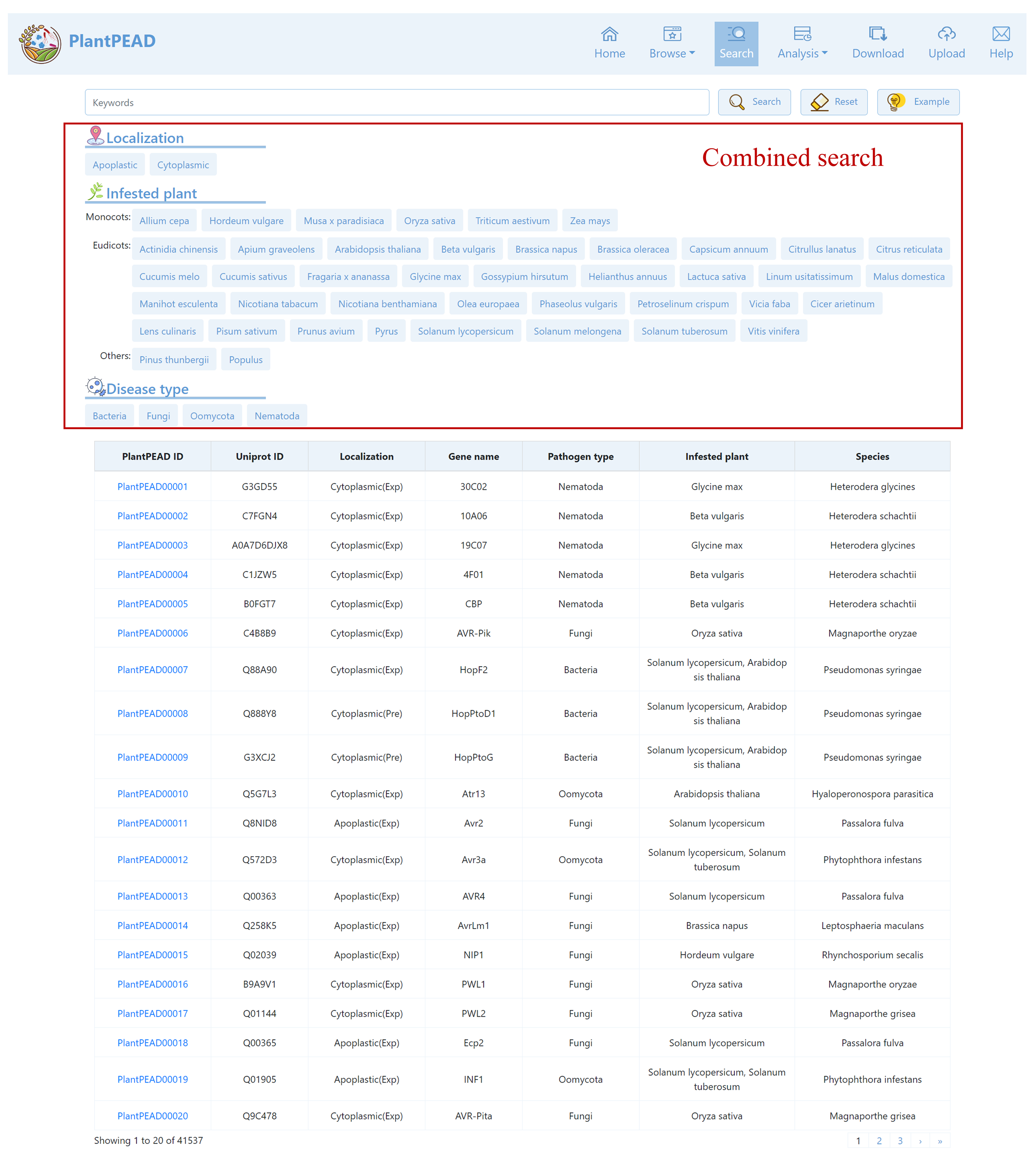
Search Page
Our search page offers a combined search feature that allows you to combine multiple searches for enhanced results.
Analysis Page
On the analysis page, five tools are provided to comprehensively explore pathogen effectors.
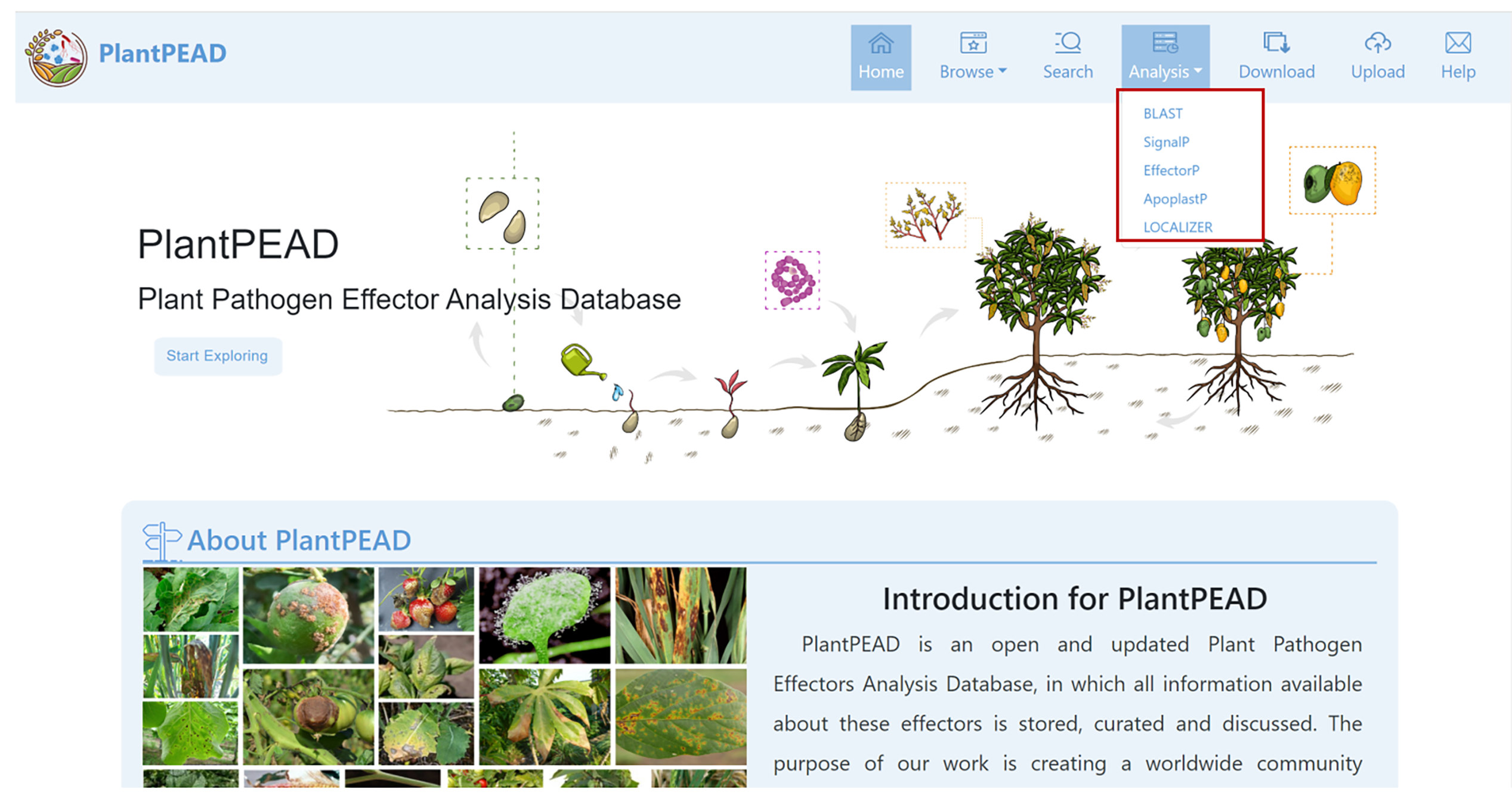
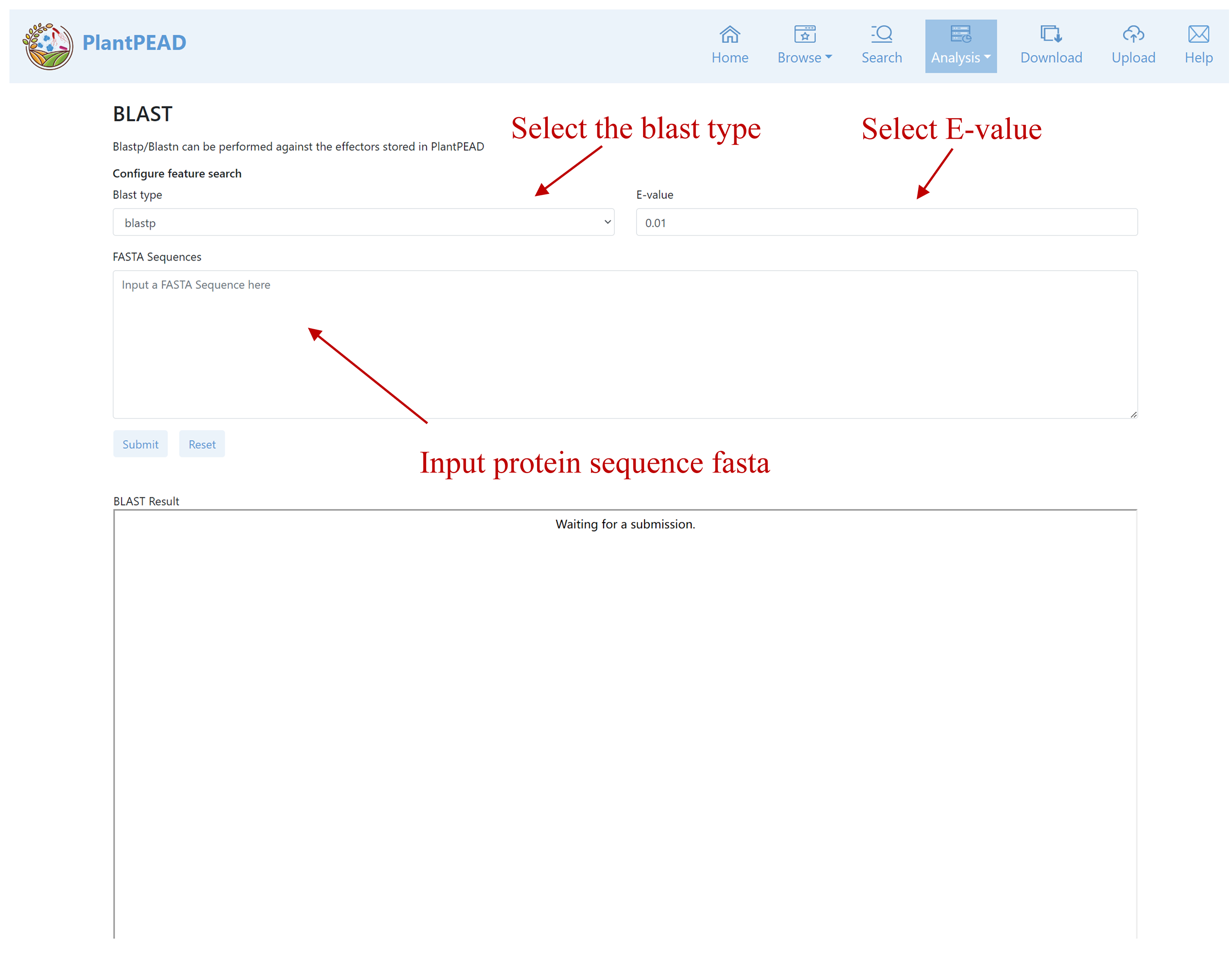
BLAST
Blastp/Blastn can be performed against the effectors stored in PlantPEAD.
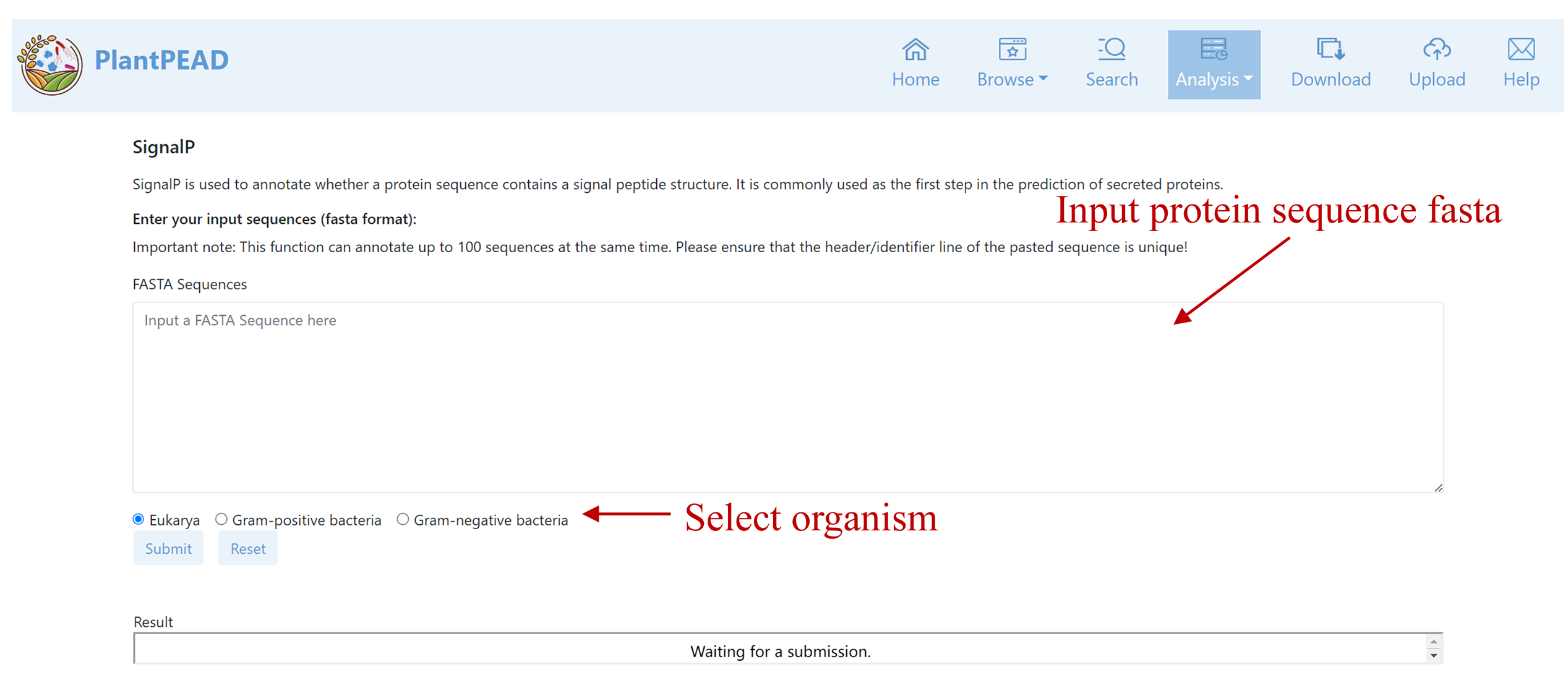
SignalP
SignalP is used to annotate whether a protein sequence contains a signal peptide structure. It is commonly used as the first step in the prediction of secreted proteins.
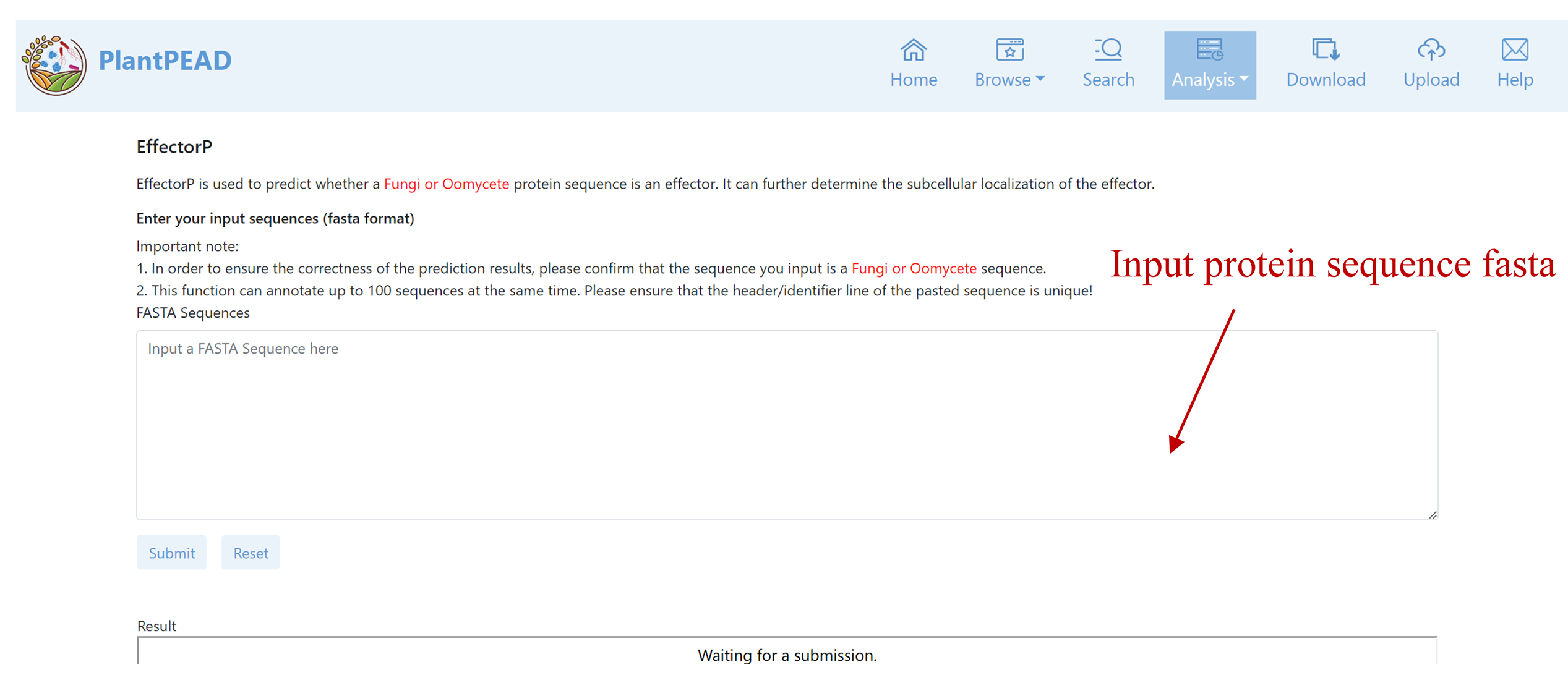
EffectorP
EffectorP is used to predict whether a Fungi or Oomycete protein sequence is an effector. It can further determine the subcellular localization of the effector.
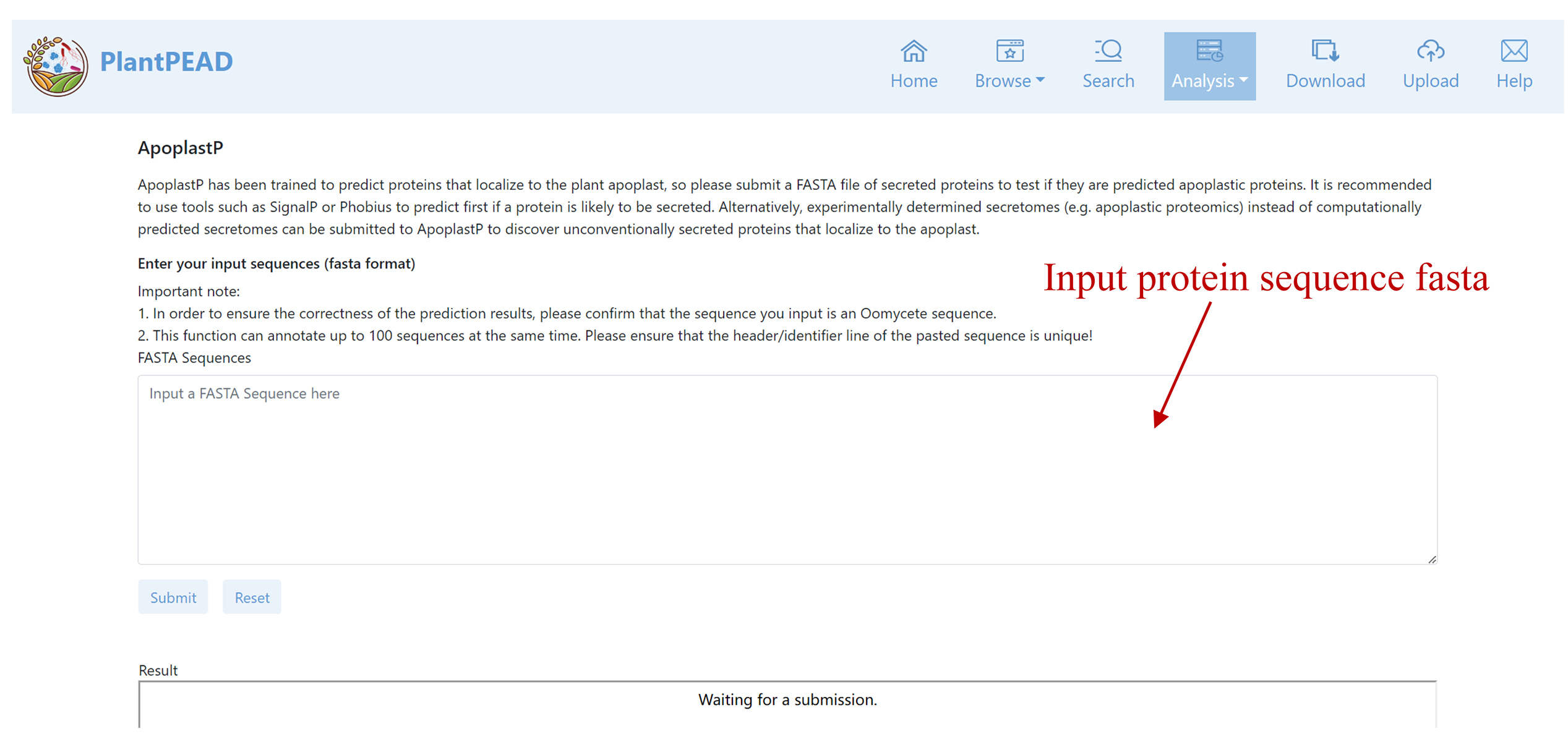
ApoplastP
ApoplastP has been trained to predict proteins that localize to the plant apoplast.
LOCALIZER
LOCALIZER is a machine learning method for predicting the subcellular localization of both plant proteins and pathogen effectors in the plant cell. It can currently predict localization to chloroplasts and mitochondria using transit peptide prediction and to nuclei using a collection of nuclear localization signals (NLSs).